
Named entity recognition (NER) is the task of recognising proper names and words from a special class in a document, such as product names, locations, people, or diseases. This can be compared to the related task of named entity linking, where the products are linked to a unique ID.
Imagine that you have received a large dataset of text in a specific domain, and would like to identify names of companies or products in the text. This could be useful if you need to identify trends such as negative sentiment towards a product in a particular context. For example, “there is a growing trend of negative sentiment around the topic iPhone”.
An industry example when you might need NER would be in the pharmaceutical industry, where pharma companies’ sales reps produce huge volumes of transcripts and notes from meetings and calls with healthcare professionals. Pharma companies are interested in insights such as “x% of mentions of a given drug by doctors are associated with a particular side effect”, which can only come from a detailed analysis of thousands of text documents.
The first stage of any analysis like this would be named entity recognition and named entity linking, whereby a system recognises the entities in question. Drugs would be identified and linked to an ID, and diseases should be linked to a particular dictionary of diseases such as MeSH terms.

Example of a named entity recognition, product recognition, or named entity linking system. The input is raw text, and the output is a tagging of all relevant entities. A named entity recogniser would tag that the 14th token is a named entity, and a named entity linker would also match it to an ID in a database.
There are a number of off-the-shelf named entity recognition libraries, such as SpaCy and the scientific domain ScispaCy, which can be used for named entity recognition. However they often over-trigger as they are trained on multi-domain texts, and you can end up with an unusable system. For example, CT is both a medical term and the state of Connecticut, and texts containing addresses can end up with a lot of false positives.
The advantage of course of using an off-the-shelf library is of course the ease of use. You can get started without needing to train a model.
Many off-the-shelf libraries for named entity recognition, such as Microsoft Azure Cognitive Services Text Analytics, or AWS Comprehend Medical, are cloud-based. This may be a consideration if your data is sensitive and confidential, as you would need to send data routinely to the cloud provider’s servers over the internet.
Depending on your use case and budget, you may find it worthwhile to build a custom made named entity recogniser.
In recent years a lot has been said and written about using deep learning for every AI problem, from natural language processing to computer vision.
However, for the above example in the pharma industry, I would actually go against the prevailing wisdom and suggest you consider a knowledge engineered approach rather than a pure machine learning approach.
It’s relatively simple to obtain a quite comprehensive list of common drug names, abbreviations, and brand names. For example, Medlineplus and the FDA both have updated lists, that you can scrape or download.
Fast Data Science - London

A sorted list of drug names that could be used in a dictionary based approach to product or named entity recognition.
You can use this and cross-check with a dictionary of common English words. Any drug name such as Minocycline that doesn’t coincide with the English dictionary can be entered in your dictionary as a definite match. Words or abbreviations in particular which do coincide with common English words can be entered in the dictionary as a low confidence match.
Since many drugs are referred to by abbreviations, you can use the dictionary approach combined with a few rules, such as allowing the last few letters to be omitted from a drug name if the result would be unambiguous.
Similarly, you can download a complete list of MeSH terms from the NIH Library of Medicine as an XML file, and remove the terms which coincide with the English dictionary.
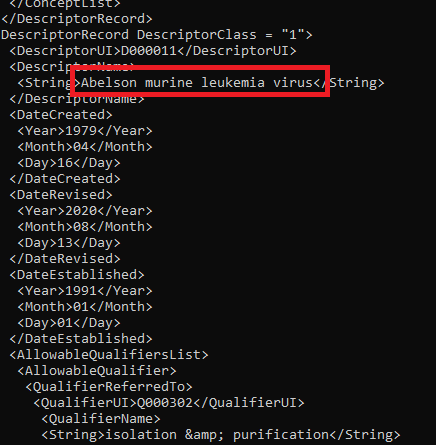
Screenshot of the MeSH terms list in XML format. A disease name is highlighted.
The above dictionary-based approach may get 95% of coverage for your use case, and you may not need to involve any machine learning at all!
We have tried this approach and have open-sourced a dictionary-based drug named entity recognition library on Github here: https://github.com/fastdatascience/drug_named_entity_recognition. It can be installed from Pypi using the command
pip install drug-named-entity-recognition
One approach to product recognition which is particularly powerful is to combine the dictionary-based approach listed above with a machine learning approach to hoover up any entities that have not been captured by the dictionary (misspellings, drugs that weren’t in the list, etc).
I would recommend a series of steps combined into an NLP pipeline as follows:
Split up the input text into words and spaces (tokenise it). I recommend to use a regex based tokeniser, which splits on word boundaries.
Apply the dictionary recogniser to all words, and mark words in the dictionary as entities.
(Optional) Apply part-of-speech tagging (mark each word as noun, verb, or adjective).
Identify a number of features of every word, such as “is all upper case”, “preceded by comma”, etc.
Based on a decision tree or trained machine learning model, or even some hand-coded rules, mark some of these words as low-confidence entities. For example, a word may occur in the input, which from its surrounding context is highly likely to be a drug name (“I routinely prescribe X to my patients”), but isn’t in the dictionary. It could be a misspelling, an unknown abbreviation, or a new drug. In these cases the model should pick it up and flag it to the user.
A standard approach to the named entity tagging problem is to use a notation called IOB (Inside-Outside-Beginning) notation. So every token is marked as Inside, Outside, or Beginning. So, if we imagine we are processing the sentence:
…treatment with dihydroartemisinin-piperaquine and primaqunine to kill malaria parasites.
then we would mark tokens with B-DRUG (beginning of a drug name), I-DRUG (inside a drug name), etc.
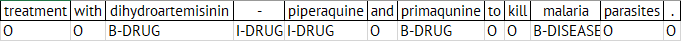
Then either you can set up hardcoded rules (“treatment with” likely to precede B-DRUG), or a machine learning model can learn some rules to apply to unseen texts.
One of the commonly used algorithms for named entity recognition which uses the IOB notation is Conditional Random Fields (CRFs). You can read more about CRFs and training a named entity recognition model in this way on the Scikit-Learn website which has a tutorial.
The advantage of the rule based approach, or even some of the simpler machine learning based approaches, is that they are relatively easy for humans to understand. In particular a knowledge engineered dictionary can be edited on demand by non-technical personnel, allowing management to understand why certain words were tagged or missed.
Sometimes a higher performance can be delivered using state of the art deep learning models.
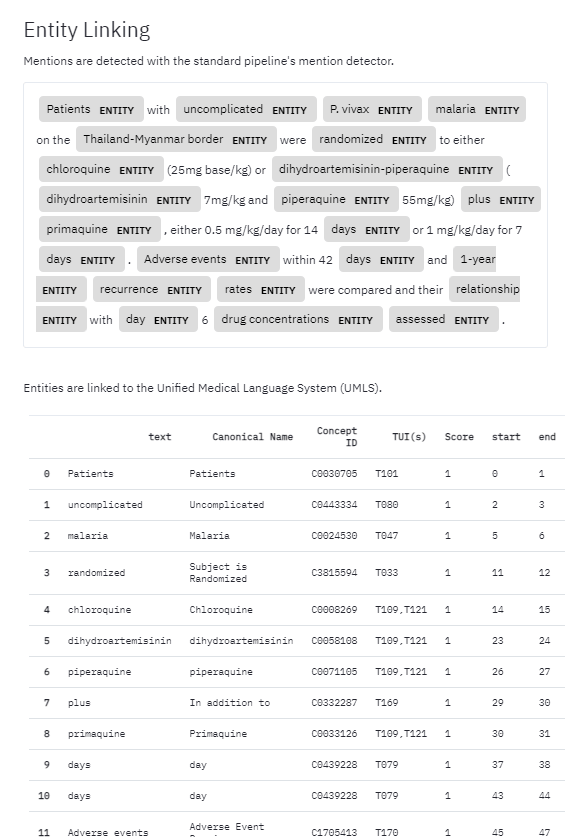
Screenshot of ScispaCy output for a sentence in the biomedical domain.
There are a number of more sophisticated named entity recognition models. A commonly used package in Python is spaCy, which uses a deep convolutional neural network to recognise entities, typically persons, organisations, and locations. A scientific domain trained model called ScispaCy is also available.
Models such as CRFs are limited to looking at small windows of text around a candidate entity, and convolutional networks also iterate a window over the document. Slightly higher performant models are Long Short Term Memory (LSTM), which is able to take into account the entire preceding context of a document.
The current state of the art is the family of transformer based neural networks such as BERT, which use an “attention mechanism” to focus on certain words in the sentence which are particularly relevant to the word in question.
The deep learning techniques for named entity recognition require large amounts of computing power, so you may need to use a cloud provider such as Azure, AWS, or Google Cloud Services, and also need a lot of tagged data. So in most cases, if you have for example a large list of drugs or molecules as an XML file which you want to pick out, then you are better off using a dictionary based approach rather than leaping right into deep learning.
Many clients or developers may feel that it’s an either-or question: do we use pure knowledge engineering, or plump for the most cutting-edge deep learning model for our product recognition task?
In fact, many NLP problems lend themselves to a hybrid approach, where independent models can be stitched together, allowing the best of both worlds.
Why not use a dictionary approach for the high-confidence unambiguous drug names, product names, or locations, combined with a machine learning based probabilistic approach for potential ambiguities?
This is what we often recommend to clients and find that in practice it works very well.
The choice of a named entity recognition algorithm therefore depends on
whether named entity recognition is needed, or whether a full named entity linking system is also needed,
the availability of a list of entities,
the availability of a tagged text dataset,
the need of the business for a transparent and accountable algorithm - or can we get away with a black box algorithm?
the need for privacy (can cloud services be used),
the number of variants of each entity - is this known? are entities likely to be misspelt?
the relative costs of false positives and false negatives returned by the recogniser.
If you have a need to recognise a large dataset of products, drugs, molecules, or similar, please get in touch with our team at Fast Data Science. We have a proven track record in delivering named entity recognition solutions and can help you to decide which of the above approaches is right for your business to recognise product names in documents.
Ready to take the next step in your NLP journey? Connect with top employers seeking talent in natural language processing. Discover your dream job!
Find Your Dream Job
We are excited to introduce the new Harmony Meta platform, which we have developed over the past year. Harmony Meta connects many of the existing study catalogues and registers.

Guest post by Jay Dugad Artificial intelligence has become one of the most talked-about forces shaping modern healthcare. Machines detecting disease, systems predicting patient deterioration, and algorithms recommending personalised treatments all once sounded like science fiction but now sit inside hospitals, research labs, and GP practices across the world.
If you are developing an application that needs to interpret free-text medical notes, you might be interested in getting the best possible performance by using OpenAI, Gemini, Claude, or another large language model. But to do that, you would need to send sensitive data, such as personal healthcare data, into the third party LLM. Is this allowed?
What we can do for you