
The pharmaceutical industry is worth over a trillion dollars and has traditionally been conservative in outlook. There is great potential for AI to transform the pharmaceutical industry and introduce huge cost savings in all stages of the business, as well as impacting financial planning with use cases such as clinical trial cost modelling with NLP. Like AI in healthcare in general, uptake of machine learning, natural language processing, and AI in pharma is just recently beginning, and already pharmaceutical companies are beginning to see large returns on the initial investment. At Fast Data Science we specialise in applying natural language processing and AI to problems in the pharmaceutical and healthcare industries. To find out more, read on or get in touch with us.
Thomas Wood demos the Clinical Trial Risk Tool, an application of NLP and AI in pharma which assists a human in analysing clinical trial protocols.
The first phase of drug development is the drug discovery phase, where candidate compounds are discovered before testing in humans. Historically, candidate medications were discovered by identifying the active ingredient in traditional remedies. For example in the 1800s chemists isolated salicylic acid from the bark of the willow tree, long used as a folk remedy to treat headaches, and this became the basis of Aspirin, which is widely used today.
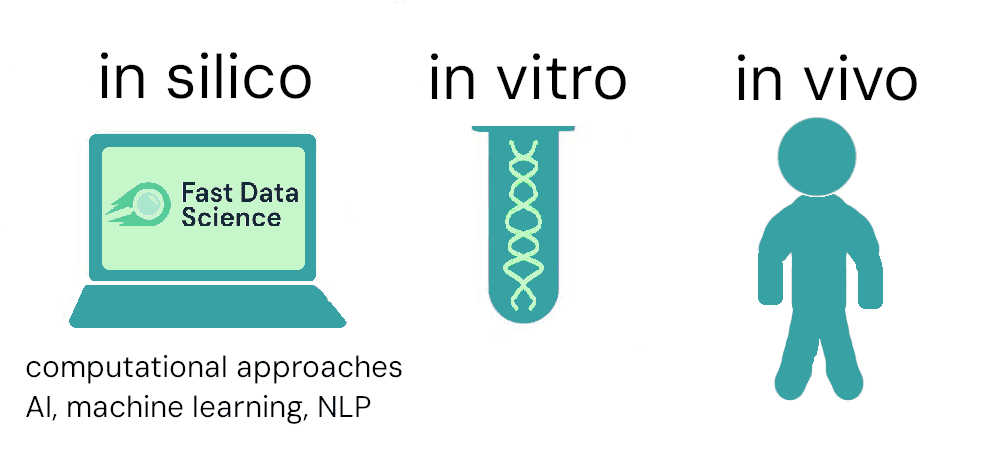
Nowadays, drug discovery involves screening millions of molecules, initially using sophisticated computer programs to simulate the interaction of the molecules with targets in the human body. This is called in silico, as a neo-Latin analogue to the in vivo - in live subjects, and in vitro - in test tubes and Petri dishes.
Before the explosion of AI use cases in pharma, researchers simulated molecule interactions using software that was pre-programmed with the rules of physics and chemistry. Recently, there has been a trend of using machine learning in pharma to learn what kind of proteins interact with what kind of targets and generalise to new unseen molecules that have not yet been synthesised.
One particularly hard problem is how to predict how a sequence of amino acids will fold into a three-dimensional structure (a protein). In 2016, researchers at DeepMind developed AlphaFold, which is able to “learn” three-dimensional shapes from databases of known proteins and predict the shape of a new sequence. The Utah-based AI/pharma company Recursion Pharmaceuticals takes AlphaFold’s predictions and tests them experimentally.
In 2015, the San Francisco-based startup Atomwise developed a convolutional neural network-based algorithm called AtomNet, which was given a dataset of observed interactions between molecules, and which was able to learn some of the rules of organic chemistry without being explicitly taught. Atomwise used AtomNet to identify lead candidates for drug research programs, and it went on to identify one candidate in the fight against the Ebola virus, which later went on to pre-clinical trials.
 [algorithms](https://harmonydata.ac.uk/measuring-the-performance-of-nlp-algorithms). Image source: Segura-Bedmar et al (2014)](https://fastdatascience.com/images/ddi-min.jpg)
AI in pharma
Some annotated texts in the DDI corpus used in the DDIExtraction Challenge, showing tagged drug-drug interactions which can be used to train machine learning algorithms in pharma. Image source: Segura-Bedmar et al (2014)
At Fast Data Science we have worked on a project for Boehringer Ingelheim, where the company has open-sourced a number of proprietary molecules. Researchers are free to order samples of the compounds in question, and the molecule structure is published online and in the literature.
The company was interested in following publications where the authors have used Boehringer’s molecules, whether or not Boehringer is cited, for the purposes of tracking any new developments arising from the discoveries, and identifying potential future collaborations. Fast Data Science developed a bespoke natural language processing algorithm to track collaborations and mentions of molecules and flag them to the drug discovery team, even if a variant of the molecule name is used.
| Pharma company | 2022 revenue |
|---|---|
| Pfizer | $100.33 billion |
| Johnson & Johnson | $94.94 billion |
| Roche | $66.26 billion |
| Merck & Co. | $59.28 billion |
| AbbVie | $58.05 billion |
| Novartis | $50.54 billion |
| Bristol Myers Squibb | $46.16 billion |
| Sanofi | $45.22 billion |
| AstraZeneca | $44.35 billion |
| GSK | $36.15 billion |
| Takeda | $30.00 billion |
| Eli Lilly | $28.55 billion |
| Gilead Sciences | $27.28 billion |
| Bayer | $26.64 billion |
| Amgen | $26.32 billion |
| Boehringer Ingelheim | $25.28 billion |
| Novo Nordisk | $25.00 billion |
| Moderna | $18.47 billion |
| Merck KGaA | $19.16 billion |
| BioNTech | $18.20 billion |
Top pharma companies worldwide by 2022 revenue. Data source
To stay competitive, pharma companies invest huge amounts in market research activities, such as interviewing doctors around their use and prescriptions of existing drugs, or trawling medical records and reports for descriptions of adverse events (AEs) even for drugs which have already passed Phase 4 trials and received FDA approval.
The pharmaceutical industry contains many smaller players which undertake market research, distribution, or other functions for the big pharma companies like those above. A number of smaller companies have contracted Fast Data Science for KOL analysis (analysis of key opinion leaders) using NLP. This can include identifying prominent academics in the field, analysing published literature, conference proceedings, or transcripts of interviews with healthcare providers. The potential for AI in ancillary functions of the pharma industry is huge.
Above: Pharmaceutical AI algorithms can be used to analyse Key Opinion Leader (KOL) insights, such as by identifying patterns in large volumes of transcripts or documents using NLP, and converting these to usable statistics which can be used for quantitative decision making.
A contract research organization (CRO) is a company that provides support to the pharma industries, for example by managing clinical trials. AI can help accelerate recruitment, and anticipate recruitment costs and attrition. CROs can analyse data to identify trends and suggest patients which are likely to benefit from a particular treatment. AI can also be used to extract data from Adverse Event Reports (AERs), clinical development plans (CDPs), and other clinical trial documents that a CRO needs to process. We’ve also worked on projects where AERs had to be anonymised to comply with GDPR or HIPAA (see our related blog post on training machine learning models on sensitive data). AI can also be used for cost analysis, such as a regression model to predict the cost or complexity of running a clinical trial.
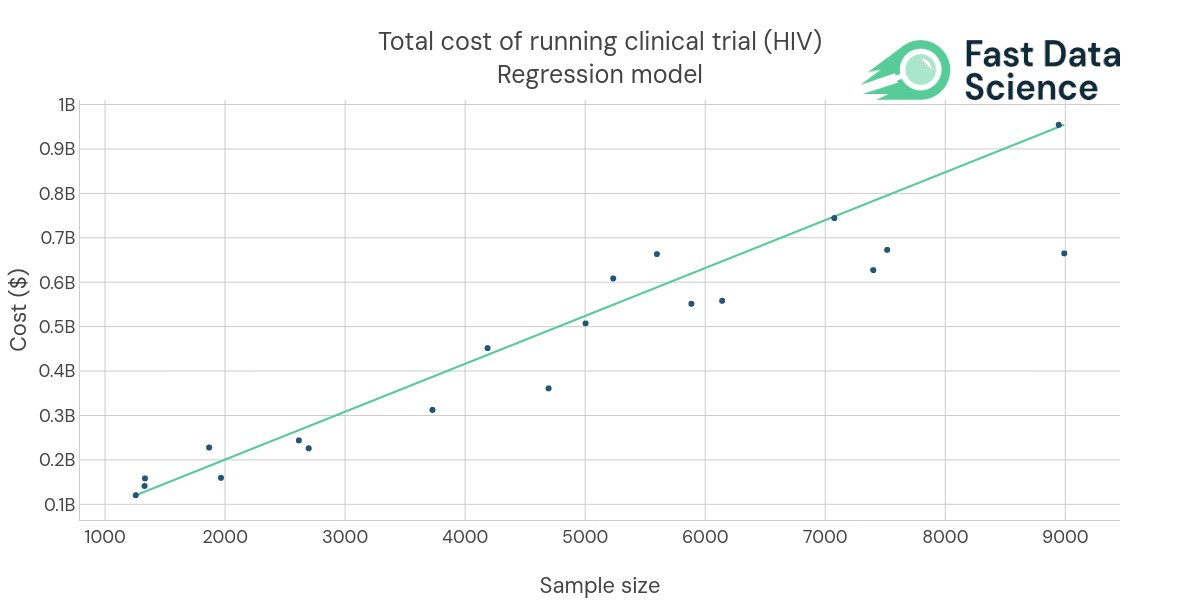
An example of one of our past pharma AI engagements: we have developed regression models for planning costs of running clinical trials. The data shown above is synthetic due to client confidentiality.
There are a number of promising companies around the world working on AI in pharma, from drug discovery and pre-clinical to clinical and after-market solutions.
| Pharma AI company | Location | Summary |
|---|---|---|
| Recursion | USA | Recursion takes the in silico AI predictions from AlphaFold and tests them experimentally. |
| Google DeepMind | UK | DeepMind first made headlines when AlphaGo defeated the world’s top human Go player, Lee Sedol of South Korea. After AlphaGo’s success, DeepMind’s charismatic founder Demis Hassabis turned his attention to 3D protein folding and produced the pharmaceutical AI program AlphaFold, which has now produced predictions of the shapes of millions of proteins. |
| Genesis Therapeutics | USA | Genesis Therapeutics has developed a generative AI drug development platform which helps pharma companies discover new medicines against molecular targets that have previously been hard for pharma companies to target. |
| Insilico Medicine | Hong Kong | Insilico uses pharma AI models to identify targets (proteins which a drug should act upon in the body) and drug candidates for diseases, such as a drug candidate, provisionally called INS018_055, which could be useful against pulmonary fibrosis and which is now in Phase 2 trials. |
| Insitro | USA | Insitro trains models on gene sequences, clinical outcomes, and pathology slides. Their models can make predictions of how cancer patients are likely to respond to particular treatments. |
| Camino | UK | Camino is using generative AI in medcomms, sifting through Pubmed abstracts and papers to find information about the pharma market such as competitors for pharma clients. |
There are many possible combinations of potential drug-drug interactions and it is a labour intensive task to read through medical literature to identify them. The danger of adverse effects from drug-drug interactions increases considerably when a patient is on multiple prescriptions.
Deep learning and text mining algorithms have been used to process the body of scientific literature and identify candidate interactions and their possible effects. Researchers developing algorithms for this purpose use the DDIExtraction Challenge as a standardised test for algorithms that identify drug-drug interactions, and every year new deep learning algorithms are improving on the top score on this metric.
Our Drug Named Entity Recognition Python library has over half a million downloads and is used around the world for analyses of free text data containing drug names. We also have a medical named entity recognition Python library.
A challenging part of drug development is the problem of identifying targets for drug development. Drug targets are molecules in the body which are associated with a particular disease. When the drug target is identified, it is possible to look for candidate molecules which are likely to interact with that target and inhibit the disease.
The candidate pancreatic cancer drug BPM31510, developed by Berg using AI drug discovery. Using this approach, Berg was able to produce a candidate drug called BPM31510, which has recently completed phase 2 trials for late-stage pancreatic cancer. The progression of the compound from in silico to in vitro was a flagship triumph for AI in pharma.
The AI biopharma company Berg has used a deep learning approach to identify drug targets. They have taken an array of tissue samples of patients with and without a particular disease, and exposed the tissues to a range of drugs and conditions. The response of the tissue is recorded and this is fed into a deep learning algorithm which searches for any likely change in the disease state, leading to candidate proteins which may be connected to the disease. The project brought the potential of AI in pharma into the public domain.
As a consulting engagement we did an analysis of a client’s factory error reports using clustering and topic detection. We were able to spot trends such as common causes of breakdowns e.g. temperature excursions, broken packaging, ventilation, etc, using NLP on the plain text of reports made by factory workers.
When candidate drugs have been identified, the pharma company will progress to clinical trials before the drug can be approved. In Phase 0 trials, the drugs are tested on a small number of people to gain some understanding of how it affects the body. Phase I trials involve giving the drug to 15 to 30 patients, to understand any side effects that may occur. This progresses through to Phase II trials, which begin to assess if the drug has an effect on the disease, and Phase III trials, which involve more than 100 patients and involve a comparison to existing drugs.
As of 2020, the average cost of bringing a new drug to market is $1.3 billion. Even then, only a small proportion of drugs in the Phase I stage make it through to Phase III and approval. When a clinical trial is run, the pharmaceutical company must write a detailed plan for the trial, typically a 200-page PDF document with sensitive information redacted, and submit it to a database such as clinicaltrials.gov.
At Fast Data Science we have developed a convolutional neural network to process clinical trial protocols for Boehringer Ingelheim and predict various complexity metrics which allow the pharma company to calculate the cost of running the trial. The neural network can read a report in plain English written by any pharma company and produce a number of quantitative metrics relating to the trial complexity. This allows the company to estimate costs in advance and plan trials to reduce both cost and risk.
Lim et al, Drug drug interaction extraction from the literature using a recursive neural network, PLoS ONE (2018)
Freedman, Hunting for New Drugs with AI, Scientific American (2020)
Segura-Bedmar et al, Lessons learnt from the DDIExtraction-2013 Shared Task (2014)
The Economist, Artificial intelligence is taking over drug development (2014)
Wouters OJ, McKee M, Luyten J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018, JAMA. 2020;323(9):844-853. doi:10.1001/jama.2020.1166
Wood, TA, A Tool To Tackle The Risk Of Uninformative Trials, Clinical Leader, 2025.
What we can do for you