
We have worked on a number of different projects where a client needed to parse scientific literature and identify occurrences of molecules or proteins.
As an example, the molecule on the right is Aspirin. This is still a trademark of Bayer in some countries. But in a paper it could appear under acetylsalicylic acid, 2-acetoxybenzenecarboxylic acid, C9H8O4, or a number of identifiers such as DB00945. There could also be identifiers that refer to other molecules, or identifiers that refer to only one version of a molecule.
Fast Data Science - London
Another example we have encountered often in clinical papers is the gene ERBB2, which is important in certain types of breast cancer. ERBB2 is also called Erb-B2 Receptor Tyrosine Kinase, HER2, HER-2 and many other names. These names often also refer to the protein expressed by the gene. Many names are similar to common English words, and are not always capitalised in text.
Because of these pathological effects, the task of identifying names of proteins, genes and molecules in scientific literature is fraught with difficulty. We have developed several tried and tested techniques to disambiguate these terms. Usually we need a number of annotated examples to start with, and we will train a machine learning model to learn from these examples and annotate new publications as they come in.
This can be deployed on the client’s servers and provide daily updates on a dashboard. This allows a client to monitor the literature in real time for publications around a particular molecule, protein or gene, or to spot trends in advance.
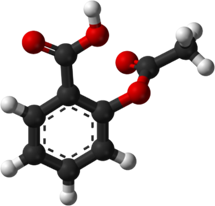
The Aspirin molecule. Source: Kim, et al, Structure Redetermination and Packing Analysis of Aspirin Crystal (1985)
What we can do for you